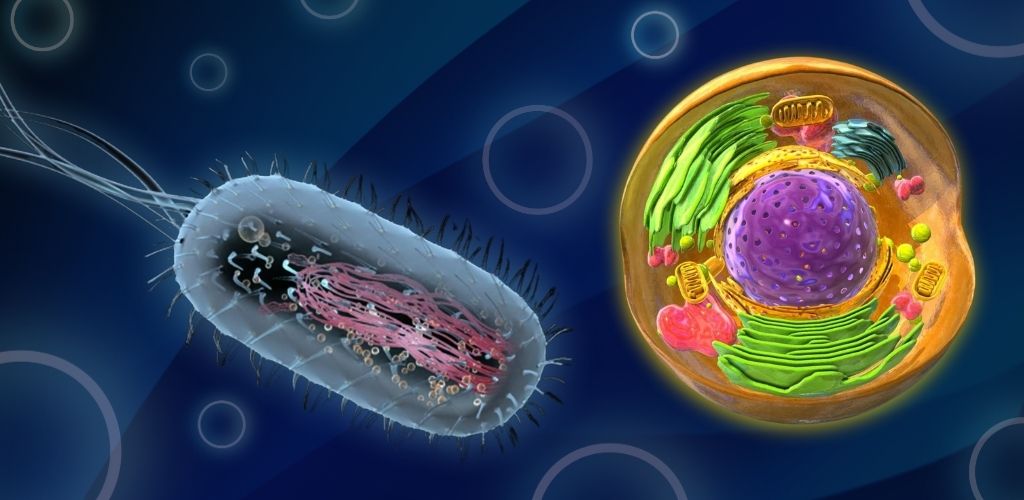
DNA packaging in prokaryotes is a distinct process from that in eukaryotes, as prokaryotic cells have a different organizational structure and lack a defined nucleus. This blog will delve into the unique aspects of DNA packaging in prokaryotes, highlighting the key differences between prokaryotic and eukaryotic cells.
DNA Packaging in Prokaryotes
In prokaryotic cells, DNA packaging is achieved through a process called supercoiling. This involves the twisting of DNA molecules, which allows them to fit within the small space of the nucleoid. The DNA in prokaryotic cells is not wrapped around histone proteins, as in eukaryotic cells, but rather relies on enzymes such as DNA gyrase to maintain its supercoiled structure.
Supercoiling in Prokaryotes
Supercoiling in prokaryotes can occur in two ways: underwinding (negative supercoiling) and overwinding (positive supercoiling). Underwinding results in a tighter coil, while overwinding creates a looser coil. The DNA’s ability to supercoil allows it to fit within the limited space of the nucleoid, which is essential for the efficient utilization of space and the maintenance of genetic stability.
Advantages of Supercoiling
The supercoiling of DNA in prokaryotes offers several advantages:
- Compactness: Supercoiling allows the DNA to fit within the small nucleoid, making it more efficient for the cell to utilize its genetic information.
- Stability: The supercoiled structure of the DNA helps maintain its stability, preventing it from degradation or damage.
- Accessibility: The supercoiled DNA can be easily accessed and read by the cell’s enzymatic machinery, allowing for the efficient expression of genes.
DNA Packaging in Eukaryotes
In contrast to prokaryotes, eukaryotic cells have a defined nucleus and employ a different packing strategy for their DNA. The DNA in eukaryotic cells is wrapped around histone proteins to form structures called nucleosomes. The histones are evolutionarily conserved proteins that are rich in basic amino acids and form a scaffold for the negatively charged DNA. This scaffold helps to organize the DNA within the nucleus, allowing it to fit within the limited space available.
Advantages of Histone-Based Packaging
The histone-based packaging of DNA in eukaryotes offers several advantages:
- Compartmentation: The histone-based packaging allows the cell to compartmentalize its genetic information, separating it from other cellular processes.
- Accessibility: The histone-based packaging makes the DNA more accessible to enzymatic machinery, allowing for the efficient expression of genes.
- Stability: The histone-based packaging helps maintain the stability of the DNA, preventing it from degradation or damage.
Comparison of DNA Packaging in Prokaryotes and Eukaryotes
While both prokaryotic and eukaryotic cells share the common goal of packaging their DNA, the methods they employ differ significantly. The key differences between the two are:
- Packing Strategy: Prokaryotes use supercoiling, while eukaryotes use histone-based packaging.
- Genome Size: Prokaryotic genomes are generally smaller than eukaryotic genomes, which may influence their packing strategies.
- Nuclear Structure: Prokaryotic cells lack a defined nucleus, while eukaryotic cells have a defined nucleus.
In conclusion, DNA packaging in prokaryotes and eukaryotes is a fundamental aspect of cell biology that allows cells to efficiently utilize and maintain their genetic information. While the methods of packaging differ between the two, both strategies serve to compact and organize the DNA, ensuring its stability and accessibility for the cell’s enzymatic machinery.
EFFECT OF DNA PACKAGING ON NGS ANALYSIS
Supercoiling’s Impact on NGS Analysis: Prokaryotes vs. Eukaryotes
NGS (Next-Generation Sequencing) is a powerful tool in genomics, but DNA’s supercoiling state can throw a wrench in the works. Here’s how it affects prokaryotes and eukaryotes differently:
Prokaryotes:
- Circular genomes: Due to their circular chromosomes, supercoiling significantly impacts prokaryotic DNA topology, affecting library preparation and sequencing efficiency.
- Densely packed: Tightly packed DNA in the nucleoid can be challenging to fragment and extract, leading to biases in sequencing coverage.
- Topoisomerase activity: Prokaryotes rely heavily on topoisomerases to manage supercoiling, their activity during sample preparation can introduce unwanted nicking or breaks, impacting read quality.
- Plasmid complexity: Plasmids present additional challenges, as their supercoiling state can differ from the chromosome and influence their representation in sequencing data.
Eukaryotes:
- Linear chromosomes: The linear nature of eukaryotic chromosomes simplifies library preparation, but supercoiling can still affect accessibility of specific regions for sequencing.
- Chromatin structure: DNA packaging with histones influences accessibility for sequencing libraries, with tightly packed nucleosomes creating sequencing biases.
- Differential supercoiling: Supercoiling levels can vary in different genomic regions and chromatin states, potentially obscuring subtle regulatory signals in NGS data.
- Nuclear organization: Spatial organization of chromosomes within the nucleus can also impact sequencing coverage, further complicating analysis.
NGS challenges and solutions:
- Library preparation optimization: Tailoring fragmentation methods and size selection can mitigate biases introduced by supercoiling.
- Computational correction: Bioinformatics tools can statistically adjust for biases stemming from supercoiling-related issues.
- Integrative analysis: Combining NGS data with other genomic measurements (e.g., Hi-C) can provide a more comprehensive picture of chromatin structure and supercoiling.
Understanding the interplay between supercoiling and NGS is crucial for accurate data interpretation in both prokaryotes and eukaryotes. By accounting for these challenges and employing appropriate techniques, researchers can unlock the full potential of NGS for deciphering the complexities of genomes.
Citations:
[1] https://courses.lumenlearning.com/wm-biology1/chapter/reading-dna-packaging-in-eukaryotes-and-prokaryotes/