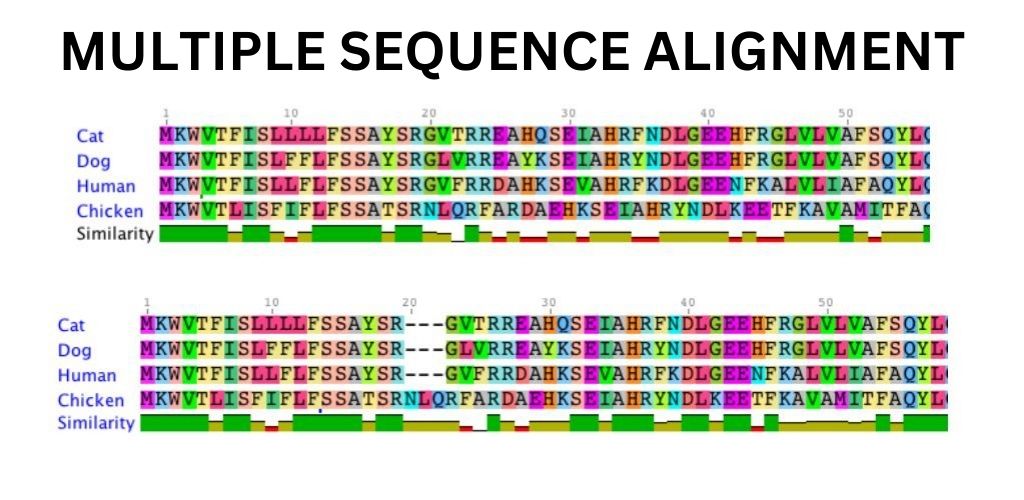
Multiple sequence alignment (MSA) is a technique used in bioinformatics to compare and analyse the similarities and differences between three or more biological sequences. The sequences can be DNA, RNA, or protein sequences. MSA is an essential tool for studying the evolutionary relationships between different species and identifying conserved regions in the sequences.
Why is MSA important?
MSA is important because it helps us to identify conserved regions in the sequences. These conserved regions are important because they are likely to have a functional significance. By identifying these regions, we can better understand the structure and function of the sequences. MSA also helps us to identify the evolutionary relationships between different species. By comparing the sequences of different species, we can determine how closely related they are.
How does MSA work?
MSA works by aligning the sequences in such a way that the maximum number of characters match. The alignment is done by inserting gaps in the sequences where necessary. The alignment is then scored based on the number of matches and mismatches. The goal is to find the alignment with the highest score.
Tools for MSA
Multiple Sequence Alignment (MSA) tools are essential in bioinformatics and computational biology for comparing and aligning three or more biological sequences, such as DNA, RNA, or protein sequences. Here’s an elaboration on the three mentioned tools: Clustal Omega, MAFFT, and Muscle.
-
Clustal Omega:
Algorithm:
Clustal Omega employs a combination of seeded guide trees and Hidden Markov Model (HMM) profile-profile techniques. The algorithm starts by creating a guide tree that represents the evolutionary relationships between sequences. It then refines the alignment using a progressive alignment strategy and profile-profile techniques.
Features:
Speed: Clustal Omega is designed to be fast and efficient, making it suitable for large-scale sequence analysis.
Accuracy: It aims to provide accurate alignments by incorporating advanced techniques like HMM profiles.
Scalability: Clustal Omega is capable of handling a large number of sequences, making it useful for high-throughput analyses.
-
MAFFT (Multiple Alignment using Fast Fourier Transform):
Algorithm:
MAFFT employs the Fast Fourier Transform (FFT) algorithm to accelerate the computation of large-scale sequence alignments. It uses a progressive alignment strategy, similar to Clustal Omega, where sequences are initially aligned in pairs and then gradually combined into the final multiple sequence alignment.
Features:
Variants: MAFFT offers several different algorithms and methods, such as FFT-NS-2 (Fast Fourier Transform – Neighbor Joining), FFT-NS-i (iterative refinement), and others. Users can choose the most appropriate method based on the characteristics of their dataset.
Accuracy and Versatility: MAFFT is known for its accuracy and versatility, providing a range of options to accommodate diverse sequence types and sizes.
-
Muscle:
Algorithm:
Muscle utilizes a progressive alignment approach to build the alignment step by step. It starts with the most similar sequences and progressively adds less similar sequences to the alignment. This process helps to avoid common pitfalls associated with global alignment algorithms.
Features:
Speed: Muscle is designed to be fast and is particularly efficient for aligning large datasets.
Sensitivity: While it may sacrifice a bit of sensitivity compared to slower methods, Muscle still aims to produce biologically meaningful alignments.
Accuracy: Muscle strikes a balance between speed and accuracy, making it suitable for a variety of applications, especially when dealing with large datasets.
Choosing the most suitable MSA tool depends on factors such as the size of the dataset, the level of accuracy required, and the computational resources available. Researchers often experiment with multiple tools to assess which one performs best for their specific analysis.
How multiple sequence alignment (MSA) is used in bioinformatics?
- Protein Structure and Function: MSA identifies conserved regions in protein sequences, aiding in predicting protein structure and understanding functional domains.
- Phylogenetic Analysis: MSA is crucial for constructing phylogenetic trees, revealing evolutionary relationships between species based on aligned DNA or protein sequences.
- Drug Discovery: MSA helps analyze drug target sequences, guiding the design of drugs that target conserved regions for increased therapeutic efficacy.
- Functional Annotation of Genomes: MSA is used to annotate functional elements in genomes, identifying conserved non-coding regions, regulatory elements, and coding sequences.
- Viral Evolution and Vaccine Design: MSA tracks viral evolution, aiding in vaccine design by identifying conserved regions in viral sequences.
- Comparative Genomics: MSA aligns entire genomes, facilitating comparative genomics to identify conserved synteny, gene order, and functional elements across species.
- Functional Genomics and Transcriptomics: MSA compares RNA sequences, contributing to functional genomics by identifying conserved elements in transcriptomic data.
Conclusion
Multiple sequence alignment is an important tool in bioinformatics. It helps us to identify conserved regions in the sequences and to determine the evolutionary relationships between different species. There are several tools available for MSA, and each tool uses a different algorithm to perform the alignment. By using these tools, we can better understand the structure and function of biological sequences. MSA is versatile, supporting various bioinformatics applications, from understanding evolution to aiding drug design and functional genomics.